Gene Duplication
Neofunctionalization?
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements.
Is gene duplication responsible for the creation and addition of new specified complexity or information which was not already present in the life form's original genetic makeup?
Evolutionary view
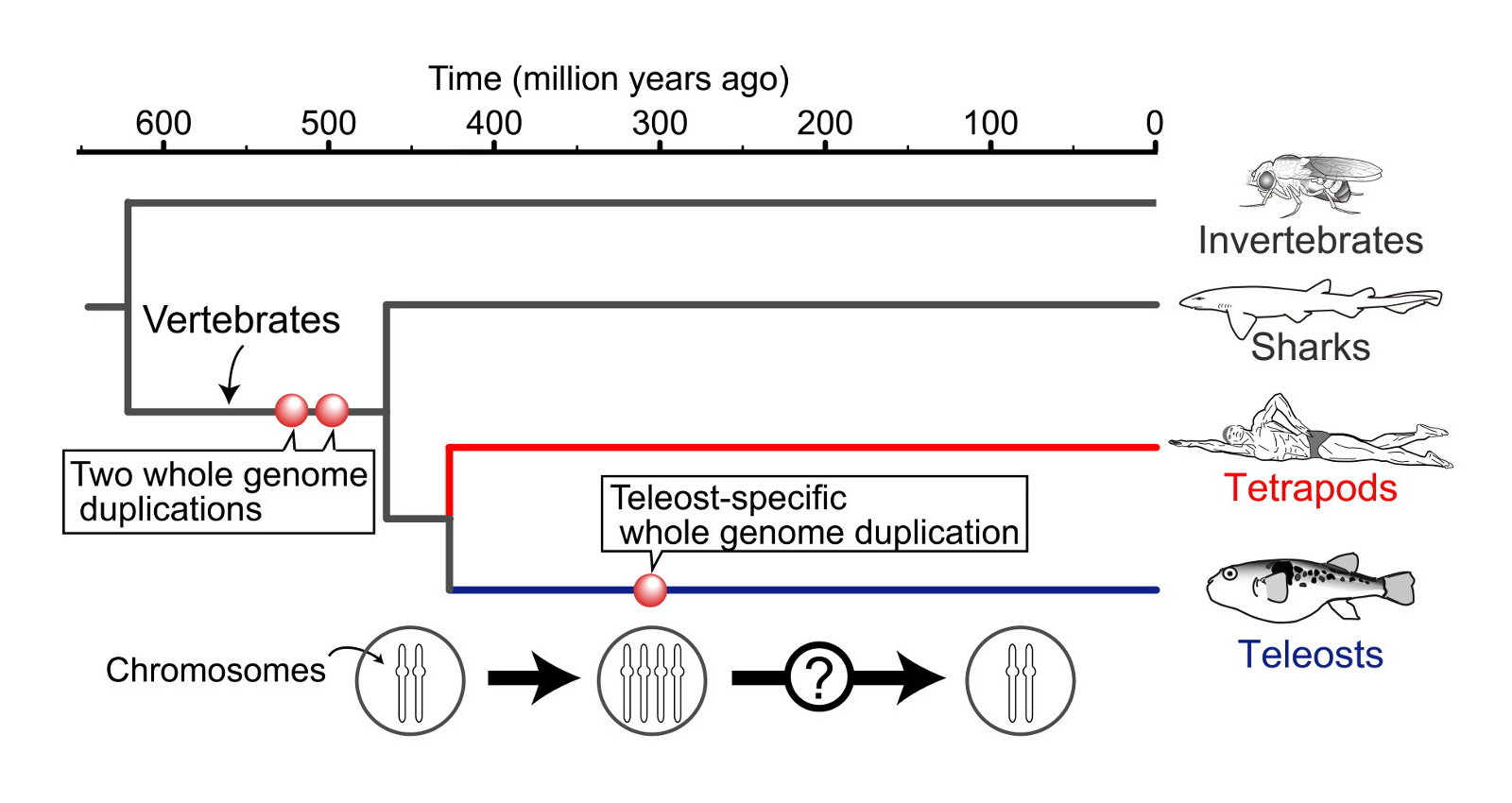
Whole genome duplication (WGD) is a rare evolutionary event that has played a dramatic role in the diversification of most eukaryotic lineages (organisms whose cells contain complex structures enclosed within membranes). Given a set of species known to have evolved from a common ancestor through one or many rounds of WGD together with a set of genome rearrangements, and a phylogenetic tree for these species, the goal is to infer the pre-duplicated ancestral genomes.
Many researchers of articles related to evolution commit circular reasoning in the sense that they assume common ancestry beforehand. They believe in macroevolution and therefore base their conclusions on that belief. This article is a perfect example of that. But of course macroevolution or common ancestry first has to be proven before one can base conclusions on it.
Gene duplication is believed to be a driving force of evolution. In his book Evolution by gene duplication, published in 1970, Susumu Ohno argued that gene duplication is the most important evolutionary force since the emergence of the universal common ancestor. Evolutionists say many things...
A gene duplication is an event in which one gene gives rise to two genes that cannot be operationally distinguished from each other. The duplicated genes remain in the same genome and therefore are paralogues and in different genome as orthologues. Gene duplication is believed to play an important role in evolution by providing material for evolution of new gene functions. A duplicated gene provides a greater, less-constrained chance for natural selection to shape a novel function.
"To shape a novel function" is every evolutionist's wet dream.
This is accomplished through a process called Gene Duplication, which is believed to play a major role in Evolution. Because of a mistake during meiosis, an organism may end up with two copies of the same gene. After this happens, the usual mechanisms of point mutation and natural selection can evolve one of the copies into coding for something completely new, while retaining the original gene.*
"Coding for something completely new" is every evolutionist's wet dream.
Gene duplication generates a copy of an existing gene. The key question here is whether it also creates new specified information which is required for the creation of new functional features which were not already present in the life form's original genetic makeup. We're looking for neofunctionalization...
Sub- or Neofunctionalization?
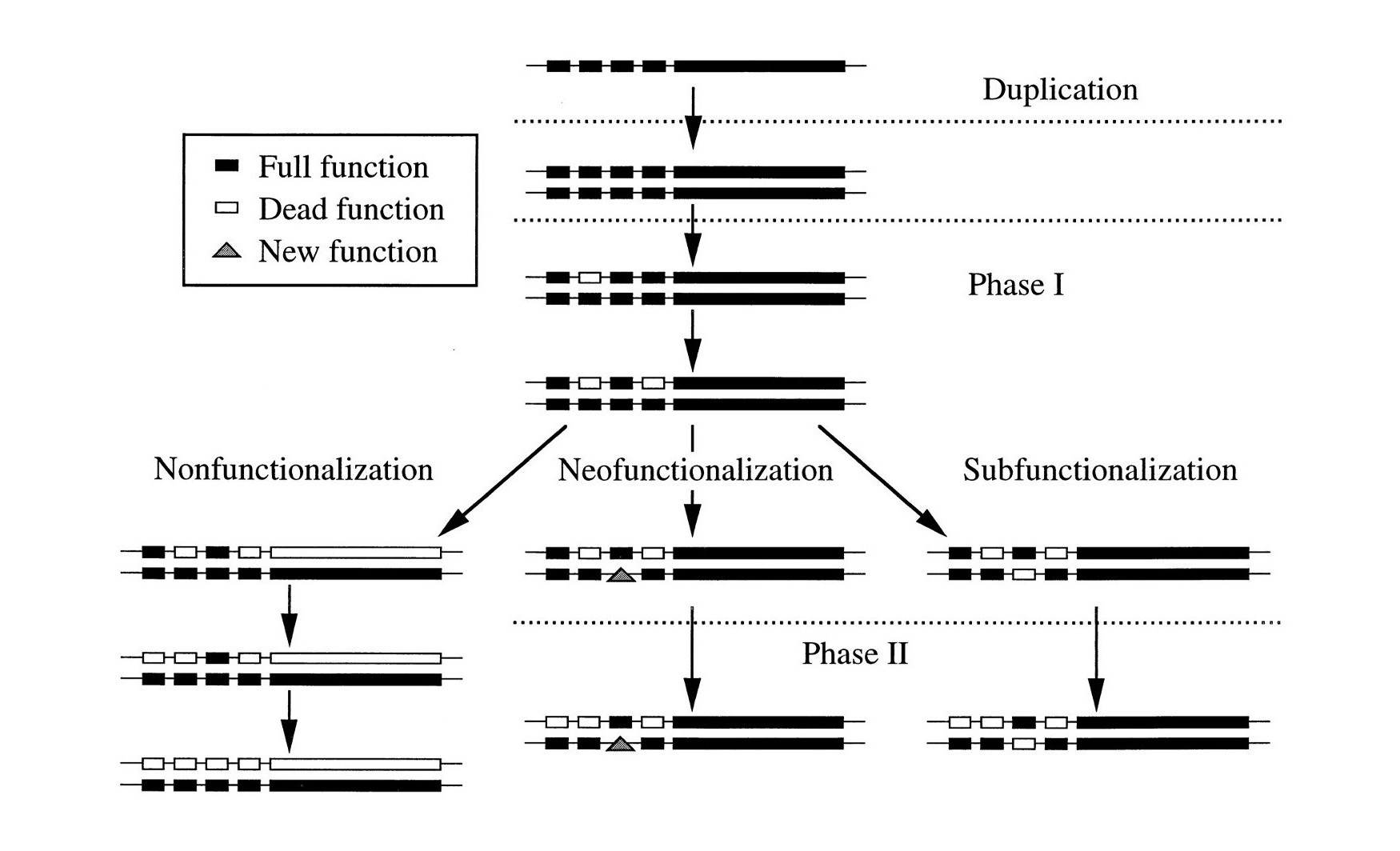
Neofunctionalization, one of the possible outcomes of functional divergence, occurs when one gene copy, or paralog, takes on a totally new function after a gene duplication event. Neofunctionalization is an adaptive mutation process; meaning one of the gene copies must mutate to develop a function that was not present in the ancestral gene.*
When a duplicated gene contains information which was already present in the original gene it is called subfunctionalization. But the question is whether completely new specified information is created and added to the new gene for macroevolution to take place. This is called neofunctionalization...
Although many of these duplicated genes might have become pseudogenes, it is possible that some acquired new functions. Identification of human-specific gene duplications might help pinpoint the genetic basis of human-unique features. With the human genome sequence now available, such studies should be feasible. In fact, several potential cases of human-specific gene duplication are known. It is also possible, as Lynch has suggested, that differential gene duplication and pseudogenization in geographically isolated populations causes reproductive isolation and speciation, although this intriguing hypothesis awaits empirical evidence.
This researcher says that most duplicated genes turn into non-functional pseudogenes and admits that evidence for speciation, or macroevolution, is still missing.
Here we present evidence that shows that the methods and models that have established neofunctionalization as a ubiquitous force in protein interaction network evolution are flawed and under reexamination support subfunctionalization, not neofunctionalization.
These researchers show that the evidence for neofunctionalization presented by other researchers is in fact merely subfunctionalization.
Gene duplication alone cannot add information to the genome. The purpose of the gene duplication in the above scenario is simply to provide raw material from which a new gene could evolve without having to give up any functions the organism already had. New information would then supposedly be built up by point mutations and natural selection. And this is precisely the process I discussed in my book and about which I said that all known examples of these mutations lose information rather than gain it.*
This was expected because in the natural world new functional information doesn't come falling from thin air. Mutations are exceedingly harmful and natural selection can do no more than prevent them from surviving. This is in line with the fact that most duplicated genes turn into non-functional pseudogenes. Pseudogenes are genomic DNA sequences similar to normal genes but non-functional; they are regarded as defunct relatives of functional genes.*
Although the process of gene duplication and subsequent random mutation has certainly contributed to the size and diversity of the genome, it is alone insufficient in explaining the origination of the highly complex information pertinent to the essential functioning of living organisms.
Degeneration
Empirical data suggest that a much greater proportion of gene duplicates is preserved than predicted by the classical model. ... The duplication-degeneration-complementation (DDC) model predicts that (1) degenerative mutations in regulatory elements can increase rather than reduce the probability of duplicate gene preservation and (2) the usual mechanism of duplicate gene preservation is the partitioning of ancestral functions rather than the evolution of new functions.
This model contains a well recognized paradox. Because duplicate genes are not under selective pressure, they should also accumulate mutations that render them incapable of encoding a protein useful for any function. Most duplicates therefore should become pseudogenes, inexpressible genetic information (junk DNA)...
Most duplicated genes turn into dysfunctional pseudogenes and they rather lose information in the process, not gain information, let alone gain new information. A copy is never better than its original. Gene duplication might be part of microevolution, variety within a species, but not of macroevolution. Gene duplication is even shown to lead to degeneration, the opposite of macroevolution. Deletion, duplication and inversion are chromosome abnormalities that have been implicated in for example autism. To believe that gene duplication is a major force of evolution is wishful thinking at best. See also Vestigiality.